Identify lung-specific transcriptomic features
2025-12-08
Last updated: 2025-12-08
Checks: 6 1
Knit directory: Lung_scMultiomics_paper/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20250512) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| ~/projects/Lung_scMultiomics_paper/data/color_codes.RDS | data/color_codes.RDS |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version b5746be. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Untracked files:
Untracked: .RData
Untracked: ArchRLogs/
Untracked: AvgHiC_ABC_three_celltypes.RDS
Untracked: Figure1.pdf
Untracked: GRNs_cross_tissueDE.png
Untracked: Lung_scMultiomics_paper.Rproj
Untracked: Rplot.png
Untracked: Rplot01.png
Untracked: Rplot02.png
Untracked: _workflowr.yml
Untracked: analysis/.ipynb_checkpoints/
Untracked: analysis/ArchRLogs/
Untracked: analysis/Figure2.R
Untracked: analysis/Figure4.R
Untracked: analysis/Figure4_ppt.R
Untracked: analysis/Figure6.R
Untracked: analysis/Figure6_ppt.R
Untracked: analysis/Plots/
Untracked: analysis/Rplots.pdf
Untracked: analysis/TF_GRN_analysis.Rmd
Untracked: analysis/TF_GRN_analysis_cross_tissue.Rmd
Untracked: analysis/TF_GRN_analysis_old.Rmd
Untracked: analysis/about.knit.md
Untracked: analysis/archive.Rmd
Untracked: analysis/dictys/
Untracked: analysis/figures_for_grant_application.Rmd
Untracked: analysis/finalize_p2g_analysis.Rmd
Untracked: analysis/finalize_p2g_analysis_30CREs.Rmd
Untracked: analysis/finalize_p2g_analysis_50CREs.Rmd
Untracked: analysis/gene.Rmd
Untracked: analysis/gene_regulatory_network_analysis.Rmd
Untracked: analysis/glmpca.Rmd
Untracked: analysis/identify_lung_specific_epigenetic_features.Rmd
Untracked: analysis/link_peaks_to_genes.Rmd
Untracked: analysis/link_peaks_to_genes_filtered.Rmd
Untracked: analysis/link_peaks_to_genes_wilcoxon.Rmd
Untracked: analysis/make_figure_panels.Rmd
Untracked: analysis/make_figures.Rmd
Untracked: analysis/make_locus_plots.Rmd
Untracked: analysis/peak_to_gene_analyses.Rmd
Untracked: analysis/peak_to_gene_analyses_50CREs.Rmd
Untracked: analysis/preprocess_scRNA_seq_data.ipynb
Untracked: analysis/publication/
Untracked: analysis/run_dimension_reduction_glmPCA.ipynb
Untracked: analysis/run_peak2gene_linkage.Rmd
Untracked: analysis/summarize_candidate_genes.Rmd
Untracked: analysis/summarize_candidate_genes.nb.html
Untracked: code/compare_tau_star_calculations.R
Untracked: code/identify_asthma_CREs.R
Untracked: code/identify_marker_peaks_edgeR.R
Untracked: code/make_plots.R
Untracked: code/plot_heatmap_asthma_CREs.R
Untracked: code/preprocess/
Untracked: code/run_GO_enrichment.R
Untracked: code/run_GO_enrichment_GRN.R
Untracked: code/run_archR_p2g_analyses.R
Untracked: code/track_plots_mapgen.R
Untracked: code/utility_function.R
Untracked: code/utils_mapgen.R
Untracked: data/AvgHiC_ABC_B_ENCODE.RDS
Untracked: data/AvgHiC_ABC_CD4_T_Corces2016.RDS
Untracked: data/AvgHiC_ABC_CD4_T_Corces2016.txt
Untracked: data/AvgHiC_ABC_CD8_T_Corces2016.RDS
Untracked: data/SCENIC_plus_TF_gene.csv
Untracked: data/aoa_ePIPs.txt
Untracked: data/aoa_gene_scores.txt
Untracked: data/asthma_GWAS_snps_hg38.RDS
Untracked: data/asthma_combined_fine_mapping_gwas_L5.rds
Untracked: data/asthma_related_CREs_combined.RDS
Untracked: data/asthma_related_CREs_combined_all_p2g_links.RDS
Untracked: data/asthma_related_CREs_combined_all_p2g_long_tbl.RDS
Untracked: data/asthma_related_CREs_combined_all_p2g_long_tbl.tsv
Untracked: data/asthma_related_CREs_combined_heatmap.RDS
Untracked: data/asthma_related_CREs_combined_heatmap_order.RDS
Untracked: data/asthma_related_CREs_combined_p2g_full.RDS
Untracked: data/asthma_related_CREs_combined_p2g_max.RDS
Untracked: data/asthma_related_CREs_full.RDS
Untracked: data/asthma_related_CREs_heatmap.RDS
Untracked: data/asthma_related_CREs_high_ePIPs.RDS
Untracked: data/asthma_related_CREs_high_ePIPs_p2g_full.RDS
Untracked: data/asthma_related_CREs_p2g.RDS
Untracked: data/asthma_related_CREs_strong_links.RDS
Untracked: data/asthma_risk_genes_two_studies.txt
Untracked: data/coa_ePIPs.txt
Untracked: data/coa_gene_scores.txt
Untracked: data/color_codes.RDS
Untracked: data/comparing_min_pct_GO.RData
Untracked: data/lung_RNA_CPM_by_CellType.RDS
Untracked: data/p2g_res/
Untracked: data/p2g_v3/
Untracked: data/pseudobulk_rna_CPM.RDS
Untracked: data/sample_covariates.txt
Untracked: data/scE2G_gene_list.txt
Untracked: data/scE2G_signif_links.RDS
Untracked: data/u19_full_atac_cell_metadata.RDS
Untracked: data/u19_full_atac_cell_metadata_with_sampleID.RDS
Untracked: keep_keys.cfg
Untracked: output/BActivationTF_targets_L_MemB.txt
Untracked: output/CD4_T_spleen_bulk_DEG.txt
Untracked: output/CD8_T_spleen_bulk_DEG.txt
Untracked: output/CD9_T_spleen_bulk_DEG.txt
Untracked: output/CEBPD_targets_L_MemB.txt
Untracked: output/CXXC1_targets_CD4_T.txt
Untracked: output/Differential_accessibility_by_celltype.xlsx
Untracked: output/Differential_accessibility_cross_tissue.xlsx
Untracked: output/Differential_expression_cross_tissue.xlsx
Untracked: output/E2F4_targets_L_MemB.txt
Untracked: output/E2G_links_vs_ethan_scores.RDS
Untracked: output/E2G_target_genes_asthma_CREs.txt
Untracked: output/EGR2_ALL_L_MemB.txt
Untracked: output/EGR2_SOX5_RUNX1_L_MemB.txt
Untracked: output/EGR2_targets_L_MemB.txt
Untracked: output/ELK1_targets_L_MemB.txt
Untracked: output/ELK4_targets_CD4_T.txt
Untracked: output/Figure1.pdf
Untracked: output/Figure1_double.pdf
Untracked: output/Figure4_double.pdf
Untracked: output/Figure6.pdf
Untracked: output/FigureS5.pdf
Untracked: output/GATA3_targets_L_MemB.txt
Untracked: output/Gene_set_enrichment_lung_upregulated_genes.xlsx
Untracked: output/Gene_set_enrichment_spleen_upregulated_genes.xlsx
Untracked: output/IRF8_targets_Th17.txt
Untracked: output/JUN_targets_Th17.txt
Untracked: output/MAF_targets_Th17.txt
Untracked: output/MAZ_targets_L_MemB.txt
Untracked: output/MEF2B_targets_L_MemB.txt
Untracked: output/MemB_cross_tissue_bulk_DEG.txt
Untracked: output/NaiveB_cross_tissue_bulk_DEG.txt
Untracked: output/Plot-UMAP-Discrepant_Genes-P2GLinks-GeneIntegrationScores.pdf
Untracked: output/Plot-UMAP-Discrepant_Genes-P2GLinks-GeneScores.pdf
Untracked: output/Plot-UMAP-E2G-GeneIntegration.pdf
Untracked: output/Plot-UMAP-E2G-GeneScores.pdf
Untracked: output/SP2_targets_CD4_T.txt
Untracked: output/TBP_targets_L_MemB.txt
Untracked: output/TF_postivie_regulators_L_MemB_DE.txt
Untracked: output/THAP1_targets_L_MemB.txt
Untracked: output/ZBTB6_ALL_L_MemB.txt
Untracked: output/all_targets_L_CD4_T_GRN.txt
Untracked: output/all_targets_L_GRN.txt
Untracked: output/all_targets_L_MemB.txt
Untracked: output/all_targets_L_Th17.txt
Untracked: output/all_targets_L_Th17_GRN.txt
Untracked: output/archR_all_genes_tested.RDS
Untracked: output/asthma_CRE_p2g_links_final_list.txt
Untracked: output/asthma_CREs_p2g_combined_tbl.txt
Untracked: output/asthma_CREs_p2g_combined_tbl_ALL.txt
Untracked: output/asthma_CREs_p2g_filtered_heatmap_inputs.RData
Untracked: output/asthma_E2G_linked_genes.txt
Untracked: output/asthma_E2G_linked_genes_full.txt
Untracked: output/asthma_candidate_risk_genes.xlsx
Untracked: output/asthma_candidate_risk_genes_full.xlsx
Untracked: output/co_activation_scE2G_multiome_tbl.txt
Untracked: output/co_activation_table_scE2G.txt
Untracked: output/co_activation_table_scE2G_gene_list.txt
Untracked: output/genomic_tracks.pdf
Untracked: output/genomic_tracks2.pdf
Untracked: output/genomic_tracks_CD4T_specific.pdf
Untracked: output/genomic_tracks_CXCR5_loci.pdf
Untracked: output/genomic_tracks_LRRC32_loci.pdf
Untracked: output/genomic_tracks_RPS25_loci.pdf
Untracked: output/genomic_tracks_not_shared_blood.pdf
Untracked: output/genomic_tracks_selected.pdf
Untracked: output/genomic_tracks_selected_loci.pdf
Untracked: output/genomic_tracks_selected_loci_80K.pdf
Untracked: output/genomic_tracks_selected_loci_scE2G.pdf
Untracked: output/lung_GRN_TFs.csv
Untracked: output/lung_GRN_regulatory_markers.csv
Untracked: output/marker_peaks_cross_tissue_DA.pdf
Untracked: output/marker_peaks_cross_tissue_DA_FOSB.pdf
Untracked: output/marker_peaks_cross_tissue_DA_HSPA1A.pdf
Untracked: output/master_regulators_GO_enrichment.RDS
Untracked: output/master_regulators_sig.RData
Untracked: output/p2g_57_links.txt
Untracked: output/p2g_links_ArchR.txt
Untracked: output/p2g_links_scE2G.txt
Untracked: output/p2g_links_scE2G_mapped_to_archR.txt
Untracked: output/p2g_links_scE2G_scATAC_mapped_to_archR.txt
Untracked: output/p2g_scE2G_scATAC_mapped_to_archR_peakset.txt
Untracked: output/scE2G_all_genes_tested.RDS
Untracked: output/scE2G_genes_vs_prior_genes.RDS
Untracked: output/scE2G_genome_wide_linked_genes.RDS
Untracked: output/smo_expression_markers_ImmuneLow.xlsx
Untracked: output/smo_expression_markers_LungAirWay.xlsx
Untracked: output/supp_Figure6_1.pdf
Untracked: output/u19_multiomics
Untracked: plots/
Untracked: ranking_genes.png
Untracked: references.bib
Untracked: references_sim.bib
Untracked: simplify_bib.sh
Untracked: tables/
Unstaged changes:
Modified: README.md
Deleted: analysis/TF_mediated_cross_tissue_DE.Rmd
Modified: analysis/TFs_drive_cross_tissue_DE.Rmd
Modified: analysis/TFs_drive_cross_tissue_DE_corrected.Rmd
Modified: analysis/heritability_enrichment_for_lung_open_chromatin.Rmd
Modified: analysis/identify_cell_types.Rmd
Modified: analysis/identify_lung_specific_transcriptomic_features.Rmd
Modified: analysis/test.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown
(analysis/identify_lung_specific_transcriptomic_features_bulk.Rmd)
and HTML
(docs/identify_lung_specific_transcriptomic_features_bulk.html)
files. If you’ve configured a remote Git repository (see
?wflow_git_remote), click on the hyperlinks in the table
below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | b5746be | Jing Gu | 2025-12-08 | identify bulk-level DE results |
Differential gene expression analyses with DESeq2
log(Counts) ~ factor(submission_date) + Race + Age + Sex + Tissue
Summarizing DE genes by selecting p-value cutoffs
A table of cell counts by tissue and cell-type.
lungs spleens
Other 1654 104
Treg 1336 47
Th17 2732 68
CD4_T 6980 886
CD8_T 12210 421
NK 8067 464
Memory_B 5287 10507
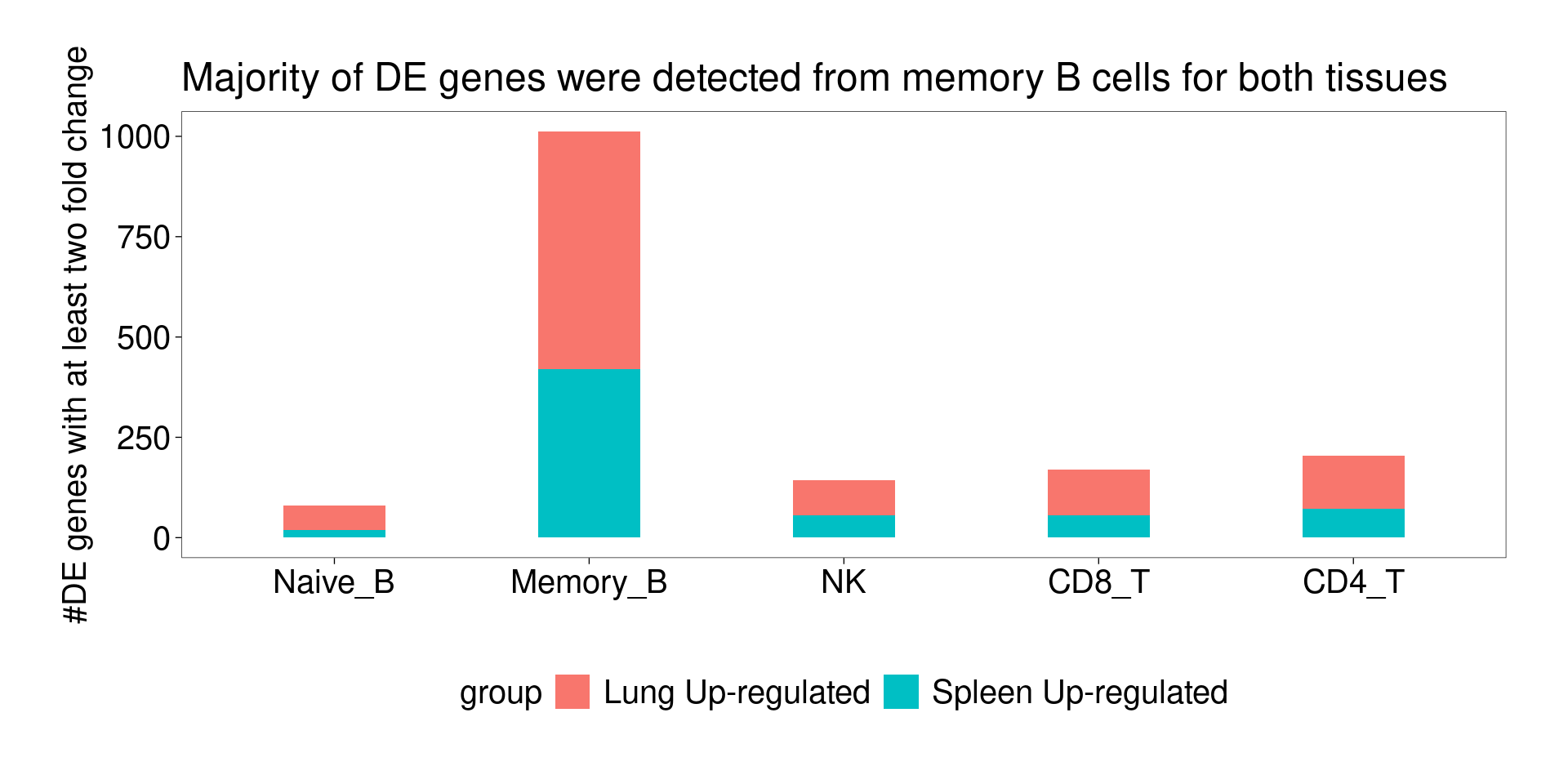
Naive_B 1174 1710A barplot for number of DE genes detected for each cell type except for Th17 and Treg, due to low number of cells in spleen.
A combined volcano plot for DE genes across cell types

DE statistics for the GNLY example
baseMean log2FoldChange lfcSE stat pvalue padj gene
1 1757.30559 2.323263 0.5248658 4.426395 9.582078e-06 9.551590e-04 GNLY
2 535.68527 6.856581 1.3146697 5.215440 1.833813e-07 1.275976e-05 GNLY
3 37.37406 4.564120 0.7593359 6.010673 1.847542e-09 1.420298e-06 GNLY
cluster avg_log2FC group
1 NK 2.323263 Lung Up-regulated
2 Memory_B 6.856581 Lung Up-regulated
3 Naive_B 4.564120 Lung Up-regulatedGO enrichment for cross-tissue genes
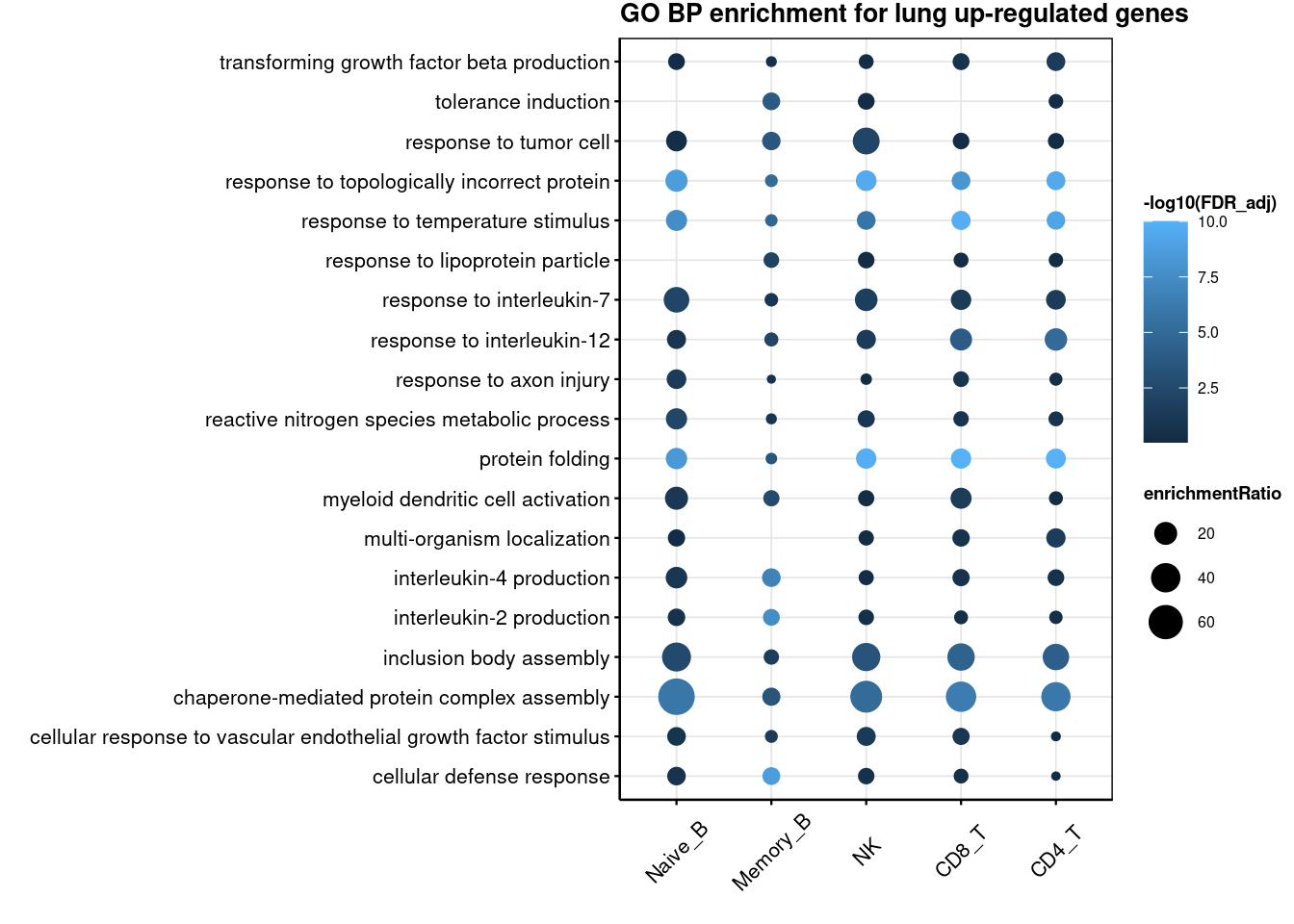
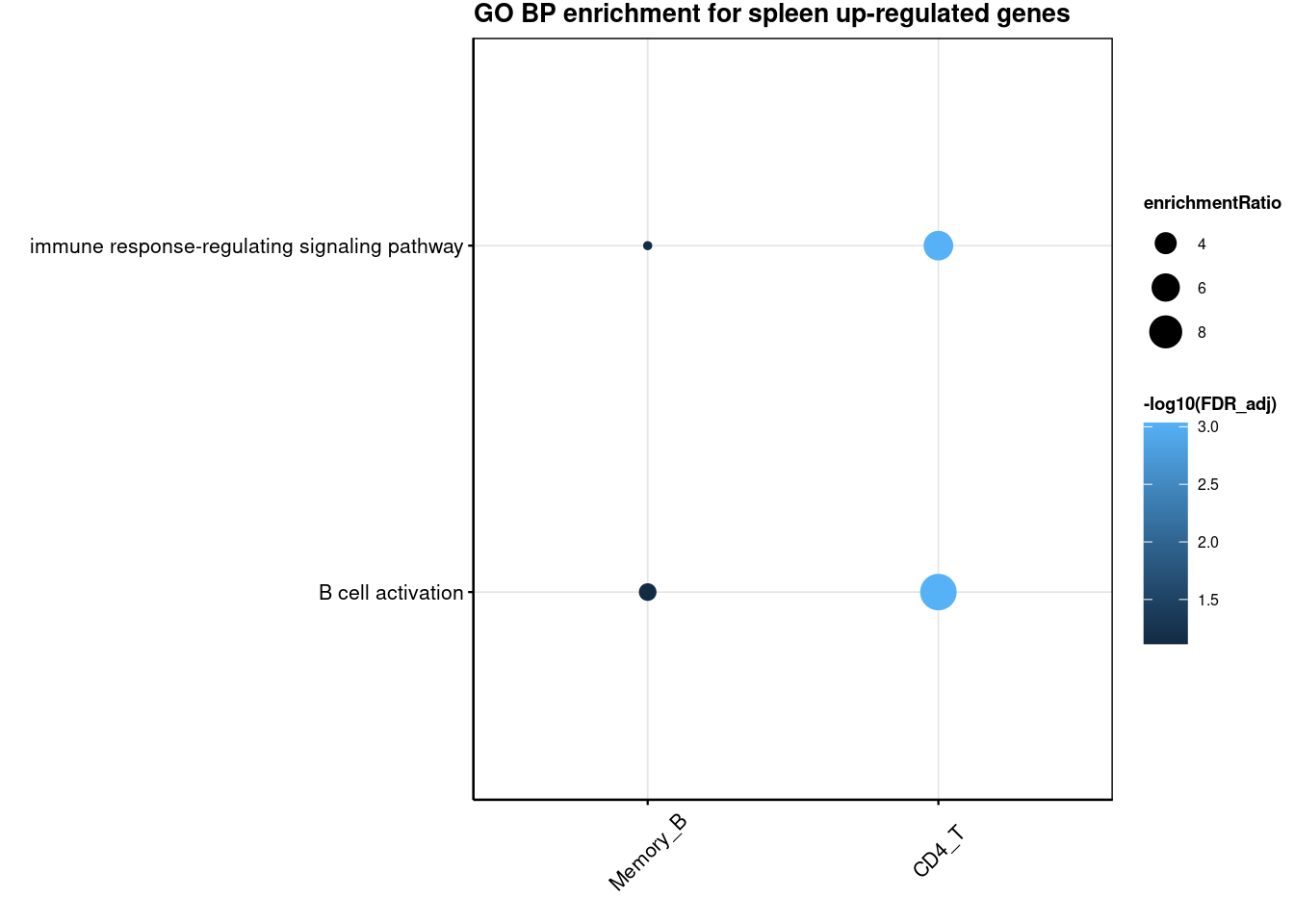
R version 4.2.0 (2022-04-22)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.3.13-el7-x86_64/lib/libopenblas_haswellp-r0.3.13.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=C
[4] LC_COLLATE=C LC_MONETARY=C LC_MESSAGES=C
[7] LC_PAPER=C LC_NAME=C LC_ADDRESS=C
[10] LC_TELEPHONE=C LC_MEASUREMENT=C LC_IDENTIFICATION=C
attached base packages:
[1] stats4 grid stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] ggrepel_0.9.6 SingleCellExperiment_1.20.1
[3] cowplot_1.1.3 ComplexHeatmap_2.14.0
[5] htmltools_0.5.8.1 scales_1.4.0
[7] colorRamp2_0.1.0 tidyr_1.3.1
[9] dplyr_1.1.4 rhdf5_2.42.1
[11] SummarizedExperiment_1.28.0 Biobase_2.58.0
[13] MatrixGenerics_1.10.0 Rcpp_1.0.14
[15] Matrix_1.6-5 GenomicRanges_1.50.2
[17] GenomeInfoDb_1.34.9 IRanges_2.32.0
[19] S4Vectors_0.36.2 BiocGenerics_0.44.0
[21] matrixStats_1.5.0 data.table_1.17.4
[23] stringr_1.5.1 plyr_1.8.9
[25] magrittr_2.0.3 ggplot2_3.5.2
[27] gtable_0.3.6 gtools_3.9.5
[29] gridExtra_2.3 ArchR_1.0.2
loaded via a namespace (and not attached):
[1] sass_0.4.10 jsonlite_2.0.0 foreach_1.5.2
[4] bslib_0.9.0 GenomeInfoDbData_1.2.9 yaml_2.3.10
[7] pillar_1.10.2 lattice_0.22-7 glue_1.8.0
[10] digest_0.6.37 RColorBrewer_1.1-3 promises_1.3.2
[13] XVector_0.38.0 colorspace_2.1-1 httpuv_1.6.16
[16] pkgconfig_2.0.3 GetoptLong_1.0.5 zlibbioc_1.44.0
[19] purrr_1.0.4 whisker_0.4.1 later_1.4.2
[22] git2r_0.33.0 tibble_3.2.1 generics_0.1.4
[25] farver_2.1.2 cachem_1.1.0 withr_3.0.2
[28] cli_3.6.5 crayon_1.5.3 evaluate_1.0.3
[31] fs_1.6.6 doParallel_1.0.17 tools_4.2.0
[34] GlobalOptions_0.1.2 lifecycle_1.0.4 Rhdf5lib_1.20.0
[37] cluster_2.1.8.1 DelayedArray_0.24.0 compiler_4.2.0
[40] jquerylib_0.1.4 rlang_1.1.6 RCurl_1.98-1.17
[43] dichromat_2.0-0.1 iterators_1.0.14 rhdf5filters_1.10.1
[46] rstudioapi_0.17.1 circlize_0.4.15 rjson_0.2.23
[49] labeling_0.4.3 bitops_1.0-9 rmarkdown_2.29
[52] codetools_0.2-20 R6_2.6.1 knitr_1.50
[55] fastmap_1.2.0 clue_0.3-66 workflowr_1.7.1
[58] rprojroot_2.0.4 shape_1.4.6 stringi_1.8.4
[61] parallel_4.2.0 vctrs_0.6.5 png_0.1-8
[64] tidyselect_1.2.1 xfun_0.52